Last Updated: February / 29 / 2024
 | About Me: |

Aaron Vose
 Resume / Curriculum Vitae. Resume / Curriculum Vitae.
 | Current Stuff: |

 | Publications: |
 Paper: Evaluating Gather and Scatter Performance on CPUs and GPUs Paper: Evaluating Gather and Scatter Performance on CPUs and GPUs
Evaluating Gather and Scatter Performance on CPUs and GPUs
Patrick Lavin, Jeffrey Young, Richard Vuduc, Jason Riedy, Aaron Vose, and Daniel Ernst
2020. The International Symposium on Memory Systems (MEMSYS 2020), Washington, DC, USA. ACM, New York, NY, USA.
|
This paper describes a new benchmark tool, Spatter, for assessing
memory system architectures in the context of a specific category
of indexed accesses known as gather and scatter. These types of
operations are increasingly used to express sparse and irregular data
access patterns, and they have widespread utility in many modern
HPC applications including scientific simulations, data mining and
analysis computations, and graph processing. However, many traditional benchmarking tools like STREAM, STRIDE, and GUPS
focus on characterizing only uniform stride or fully random accesses
despite evidence that modern applications use varied sets of more
complex access patterns.
Spatter is an open-source benchmark that provides a tunable and
configurable framework to benchmark a variety of indexed access
patterns, including variations of gather / scatter that are seen in HPC
mini-apps evaluated in this work. The design of Spatter includes
backends for OpenMP and CUDA, and experiments show how it can
be used to evaluate 1) uniform access patterns for CPU and GPU,
2) prefetching regimes for gather / scatter, 3) compiler implementations of vectorization for gather / scatter, and 4) trace-driven "proxy
patterns" that reflect the patterns found in multiple applications. The
results from Spatter experiments show, for instance, that GPUs typically outperform CPUs for these operations in absolute bandwidth
but not fraction of peak bandwidth, and that Spatter can better represent the performance of some cache-dependent mini-apps than
traditional STREAM bandwidth measurements.
|
Download:  p220-lavin.pdf p220-lavin.pdf
|
 Paper: PharML.Bind: Pharmacologic Machine Learning for Protein-Ligand Interactions Paper: PharML.Bind: Pharmacologic Machine Learning for Protein-Ligand Interactions
PharML.Bind: Pharmacologic Machine Learning for Protein-Ligand Interactions
Aaron D. Vose, Jacob Balma, Damon Farnsworth, Kaylie Anderson, and Yuri K. Peterson
2019. arXiv.org 1911.06105
|
Is it feasible to create an analysis paradigm that can analyze and then accurately and quickly predict known drugs from experimental data? PharML.Bind is a machine learning toolkit which is able to accomplish this feat. Utilizing deep neural networks and big data, PharML.Bind correlates experimentally-derived drug affinities and protein-ligand X-ray structures to create novel predictions. The utility of PharML.Bind is in its application as a rapid, accurate, and robust prediction platform for discovery and personalized medicine. This paper demonstrates that graph neural networks (GNNs) can be trained to screen hundreds of thousands of compounds against thousands of targets in minutes, a vastly shorter time than previous approaches. This manuscript presents results from training and testing using the entirety of BindingDB after cleaning; this includes a test set with 19,708 X-ray structures and 247,633 drugs, leading to 2,708,151 unique protein-ligand pairings. PharML.Bind achieves a prodigious 98.3% accuracy on this test set in under 25 minutes. PharML.Bind is premised on the following key principles: 1) speed and a high enrichment factor per unit compute time, provided by high-quality training data combined with a novel GNN architecture and use of high-performance computing resources, 2) the ability to generalize to proteins and drugs outside of the training set, including those with unknown active sites, through the use of an active-site-agnostic GNN mapping, and 3) the ability to be easily integrated as a component of increasingly-complex prediction and analysis pipelines. PharML.Bind represents a timely and practical approach to leverage the power of machine learning to efficiently analyze and predict drug action on any practical scale and will provide utility in a variety of discovery and medical applications.
|
Download:  pharml.pdf pharml.pdf
|
 Paper: Recombination of Artificial Neural Networks Paper: Recombination of Artificial Neural Networks
Recombination of Artificial Neural Networks
Aaron Vose, Jacob Balma, Alex Heye, Alessandro Rigazzi, Charles Siegel, Diana Moise, Benjamin Robbins, Rangan Sukumar
2019. arXiv.org 1901.03900
|
We propose a genetic algorithm (GA) for hyperparameter optimization of artificial neural networks which includes chromosomal crossover as well as a decoupling of parameters (i.e., weights and biases) from hyperparameters (e.g., learning rate, weight decay, and dropout) during sexual reproduction. Children are produced from three parents; two contributing hyperparameters and one contributing the parameters. Our version of population-based training (PBT) combines traditional gradient-based approaches such as stochastic gradient descent (SGD) with our GA to optimize both parameters and hyperparameters across SGD epochs. Our improvements over traditional PBT provide an increased speed of adaptation and a greater ability to shed deleterious genes from the population. Our methods improve final accuracy as well as time to fixed accuracy on a wide range of deep neural network architectures including convolutional neural networks, recurrent neural networks, dense neural networks, and capsule networks.
|
Download:  cray-hpo.pdf cray-hpo.pdf
|
 Paper: Interactive Distributed Deep Learning with Jupyter Notebooks Paper: Interactive Distributed Deep Learning with Jupyter Notebooks
Interactive Distributed Deep Learning with Jupyter Notebooks
Steve Farrell, Aaron Vose, Oliver Evans, Matthew Henderson, Shreyas Cholia, Fernando Perez, Wahid Bhimji, Shane Canon, Rollin Thomas, and Prabhat
2018, June. In International Conference on High Performance Computing (pp. 678-687). Springer, Cham.
|
Deep learning researchers are increasingly using Jupyter notebooks to implement interactive, reproducible workflows with embedded visualization, steering and documentation. Such solutions are typically deployed on small-scale (e.g. single server) computing systems. However, as the sizes and complexities of datasets and associated neural network models increase, high-performance distributed systems become important for training and evaluating models in a feasible amount of time. In this paper we describe our vision for Jupyter notebook solutions to deploy deep learning workloads onto high-performance computing systems. We demonstrate the effectiveness of notebooks for distributed training and hyper-parameter optimization of deep neural networks with efficient, scalable backends.
|
Download:  ihpc-interactive.pdf ihpc-interactive.pdf
|
 Cray Blog: How to Program a Supercomputer (October 24, 2017) Cray Blog: How to Program a Supercomputer (October 24, 2017)
 Cray Blog: The Intersection of Machine Learning and High-Performance Computing (August 31, 2017) Cray Blog: The Intersection of Machine Learning and High-Performance Computing (August 31, 2017)
 Paper: An MPI/OpenACC implementation of a high-order electromagnetics solver with GPUDirect communication. Paper: An MPI/OpenACC implementation of a high-order electromagnetics solver with GPUDirect communication.
An MPI/OpenACC implementation of a high-order electromagnetics solver with GPUDirect communication.
Otten, M., Gong, J., Mametjanov, A., Vose, A., Levesque, J., Fischer, P. and Min, M.
2016. International Journal of High Performance Computing Applications.
|
We present performance results and an analysis of a message passing interface (MPI)/OpenACC implementation of an electromagnetic solver based on a spectral-element discontinuous Galerkin discretization of the time-dependent Maxwell equations. The OpenACC implementation covers all solution routines, including a highly tuned element-by-element operator evaluation and a GPUDirect gather-scatter kernel to effect nearest neighbor flux exchanges. Modifications are designed to make effective use of vectorization, streaming, and data management. Performance results using up to 16,384 graphics processing units of the Cray XK7 supercomputer Titan show more than 2.5x speedup over central processing unit-only performance on the same number of nodes (262,144 MPI ranks) for problem sizes of up to 6.9 billion grid points. We discuss performance-enhancement strategies and the overall potential of GPU-based computing for this class of problems.
|
Download:  nekcem.pdf nekcem.pdf
|
 Paper: Massively parallel and linear-scaling algorithm for second-order Moller-Plesset perturbation theory applied to the study of supramolecular wires. Paper: Massively parallel and linear-scaling algorithm for second-order Moller-Plesset perturbation theory applied to the study of supramolecular wires.
Massively parallel and linear-scaling algorithm for second-order Moller-Plesset perturbation theory applied to the study of supramolecular wires.
Kjaergaard, T., Baudin, P., Bykov, D., Eriksen, J.J., Ettenhuber, P., Kristensen, K., Larkin, J., Liakh, D., Pawlowski, F., Vose, A., and Wang, Y.M.
2016. Computer Physics Communications.
|
We present a scalable cross-platform hybrid MPI/OpenMP/OpenACC implementation of the Divide-Expand-Consolidate (DEC) formalism with portable performance on heterogeneous HPC architectures. The Divide-Expand-Consolidate formalism is designed to reduce the steep computational scaling of conventional many-body methods employed in electronic structure theory to linear scaling, while providing a simple mechanism for controlling the error introduced by this approximation. Our massively parallel implementation of this general scheme has three levels of parallelism, being a hybrid of the loosely coupled task-based parallelization approach and the conventional MPI+X programming model, where X is either OpenMP or OpenACC. We demonstrate strong and weak scalability of this implementation on heterogeneous HPC systems, namely on the GPU-based Cray XK7 Titan supercomputer at the Oak Ridge National Laboratory. Using the "resolution of the identity second-order Moller-Plesset perturbation theory" (RI-MP2) as the physical model for simulating correlated electron motion, the linear-scaling DEC implementation is applied to 1-aza-adamantane-trione (AAT) supramolecular wires containing up to 40 monomers (2440 atoms, 6800 correlated electrons, 24440 basis functions and 91280 auxiliary functions). This represents the largest molecular system treated at the MP2 level of theory, demonstrating an efficient removal of the scaling wall pertinent to conventional quantum many-body methods.
|
Download:  lsdalton.pdf lsdalton.pdf
|
 Paper: Relative debugging for a highly parallel hybrid computer system. Paper: Relative debugging for a highly parallel hybrid computer system.
Relative debugging for a highly parallel hybrid computer system.
Luiz DeRose, Andrew Gontarek, Aaron Vose, Robert Moench, David Abramson, Minh Ngoc Dinh, Chao Jin.
2015. Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis.
|
Relative debugging traces software errors by comparing two
executions of a program concurrently - one code being a reference
version and the other faulty. Relative debugging is particularly
effective when code is migrated from one platform to another, and
this is of significant interest for hybrid computer architectures
containing CPUs accelerators or coprocessors. In this paper we
extend relative debugging to support porting stencil computation
on a hybrid computer. We describe a generic data model that
allows programmers to examine the global state across different
types of applications, including MPI/OpenMP, MPI/OpenACC,
and UPC programs. We present case studies using a hybrid
version of particle simulation DELTA5D, on
Titan at ORNL, and the UPC version of Shallow Water Equations
on Crystal, an internal supercomputer of Cray. These case studies
used up to 5,120 GPUs and 32,768 CPU cores to illustrate that the
debugger is effective and practical.
|
Download:  ccdb_pap161s4-file1.pdf ccdb_pap161s4-file1.pdf
|
 Paper: A case study of CUDA FORTRAN and OpenACC for an atmospheric climate kernel. Paper: A case study of CUDA FORTRAN and OpenACC for an atmospheric climate kernel.
A case study of CUDA FORTRAN and OpenACC for an atmospheric climate kernel
Matthew Norman, Jeffrey Larkin, Aaron Vose, and Katherine Evans
2015 Journal of Computational Science (Volume 9, Pages 1-6)
|
The porting of a key kernel in the tracer advection routines of the
Community Atmosphere Model, Spectral Element (CAM-SE), to use Graphics
Processing Units (GPUs) using OpenACC is considered in comparison to
an existing CUDA FORTRAN port. The development of the OpenACC kernel
for GPUs was substantially simpler than that of the CUDA port. Also,
OpenACC performance was about 1.5x slower than the optimized CUDA
version. Particular focus is given to compiler maturity regarding
OpenACC implementation for modern FORTRAN, and it is found that the
Cray implementation is currently more mature than the PGI
implementation. Still, for the case that ran successfully on PGI, the
PGI OpenACC runtime was slightly faster than Cray. The results show
encouraging performance for OpenACC implementation compared to CUDA
while also exposing some issues that may be necessary before the
implementations are suitable for porting all of CAM-SE. Most notable
are that GPU shared memory should be used by future OpenACC
implementations and that derived type support should be expanded.
|
Download:  cam-se.pdf cam-se.pdf
|
 Cray Blog: HPC Software Optimization Trends (Part 1): Where Did We Come From and Where Are We Now? (August 11, 2015) Cray Blog: HPC Software Optimization Trends (Part 1): Where Did We Come From and Where Are We Now? (August 11, 2015)
 Cray Blog: HPC Software Optimization Trends (Part 2): What Will the Future Be Like and How Do We Prepare for It? (August 12, 2015) Cray Blog: HPC Software Optimization Trends (Part 2): What Will the Future Be Like and How Do We Prepare for It? (August 12, 2015)
 Paper: Tri-Hybrid Computational Fluid Dynamics on DoE's Cray XK7, Titan. Paper: Tri-Hybrid Computational Fluid Dynamics on DoE's Cray XK7, Titan.
Tri-Hybrid Computational Fluid Dynamics on DoE's Cray XK7, Titan
Aaron Vose, Brian Mitchell, and John Levesque
2014 Cray User Group (CUG)
|
A tri-hybrid port of General Electric's in-house, 3D, Computational Fluid
Dynamics (CFD) code TACOMA is created utilizing MPI, OpenMP, and
OpenACC technologies. This new port targets improved performance on
NVidia Kepler accelerator GPUs, such as those installed in the second
largest supercomputer, Titan, the Department of Energy's 27 petaFLOP
Cray XK7 located at Oak Ridge National Laboratory. We demonstrate a
1.4x speed improvement on Titan when the GPU accelerators are
enabled. We highlight key optimizations and techniques used to achieve
these results. These optimizations enable larger and more accurate
simulations than were previously possible with TACOMA, which not only
improves GE's ability to create higher performing turbomachinery blade
rows, but also provides "lessons learned" which can be applied to the process of
optimizing other codes to take advantage of tri-hybrid technology with
MPI, OpenMP, and OpenACC.
|
Download:  cug2014.pdf cug2014.pdf
|
 Paper: HSPp-BLAST: Highly Scalable Parallel PSI-BLAST for Very Large-scale Sequence Searches Paper: HSPp-BLAST: Highly Scalable Parallel PSI-BLAST for Very Large-scale Sequence Searches
HSPp-BLAST: Highly Scalable Parallel PSI-BLAST for Very Large-scale Sequence Searches
Bhanu Rekepalli, Aaron Vose, and Paul Giblock
2012 Bioinformatics and Computational Biology (BICoB), ISCA 4th Int'l. Conference
|
Based on recent published articles, the
growth of genomic data has overtaken and outpaced both
performance improvements of storage technologies and
processing power due to the revolutionary advancements
of next generation sequencing technologies. By bringing
down the costs and increasing throughput by many orders
of magnitude with sequencing technologies, data is
doubling every 9 months resulting in the exponential
growth of genomic data in recent years. However, data
analysis becomes increasingly difficult and can be
prohibitive, as existing bioinformatics tools developed in
the past decade focus mainly on desktops, workstations
and small clusters that have limited capabilities.
Improving the performance and scalability of such tools
is critical to transforming ever-growing raw genomic data
into biological knowledge containing invaluable
information directly related to human health. This paper
describes a new software application which includes
optimization techniques improving the scalability of a
most widely used bioinformatics tool "PSI-BLAST" on
advanced parallel architectures, pushing the envelope of
biological data analysis. We show that our improvements
allow near-linear scaling to tens of thousands of
processing cores, up to the maximum non-capability size
on current petaflop supercomputers. This new tool
increases by 5 orders of magnitude the amount of
genomics data that can be processed per hour.
|
Download:  hspp-blast.pdf hspp-blast.pdf
|
 Paper: Patterns of Species Ranges, Speciation, and Extinction Paper: Patterns of Species Ranges, Speciation, and Extinction
Patterns of Species Ranges, Speciation, and Extinction
Aysegul Birand, Aaron Vose, and Sergey Gavrilets
2012 The American Naturalist, The University of Chicago Press
|
The exact nature of the relationship among species range sizes,
speciation, and extinction events is not well understood. The factors
that promote larger ranges, such as broad niche widths and high
dispersal abilities, could increase the likelihood of encountering new
habitats but also prevent local adaptation due to high gene flow.
Similarly, low dispersal abilities or narrower niche widths could
cause populations to be isolated, but such populations may lack
advantageous mutations due to low population sizes. Here we present a
large-scale, spatially explicit, individual-based model addressing the
relationships between species ranges, speciation, and extinction. We
followed the evolutionary dynamics of hundreds of thousands of diploid
individuals for 200,000 generations. Individuals adapted to multiple
resources and formed ecological species in a multidimensional trait
space. These species varied in niche widths, and we observed the
coexistence of generalists and specialists on a few resources. Our
model shows that species ranges correlate with dispersal abilities but
do not change with the strength of fitness trade-offs; however, high
dispersal abilities and low resource utilization costs, which favored
broad niche widths, have a strong negative effect on speciation
rates. An unexpected result of our model is the strong effect of
underlying resource distributions on speciation: in highly fragmented
landscapes, speciation rates are reduced.
|
Download:  ranges.pdf ranges.pdf
|
 Paper: A Pragmatic Approach to Improving the Large-scale Parallel I/O Performance of Scientific Applications. Paper: A Pragmatic Approach to Improving the Large-scale Parallel I/O Performance of Scientific Applications.
A Pragmatic Approach to Improving the Large-scale Parallel I/O Performance of Scientific Applications
L. D. Crosby, R. G. Brook, B. Rekepalli, M. Sekachev, A. Vose, and K. Wong
2011 Cray User Group (CUG)
|
I/O performance in scientific applications is an often neglected area of concern during performance
optimizations. However, various scientific applications have been identified which benefit from I/O improvements
due to the volume of data or number of compute processes utilized. This work details the I/O patterns and data
layouts of real scientific applications, discusses their impacts, and demonstrates pragmatic approaches to improve
I/O performance.
|
Download:  cug2011.pdf cug2011.pdf
|
 Paper: Simulating Population Genetics on the XT5 Paper: Simulating Population Genetics on the XT5
Simulating Population Genetics on the XT5
Duenez-Guzman, A. Vose, M. Vose, Gavrilets
Cray User Group 2009, Compute the Future
|
We describe our experience developing custom C code for
simulating evolution and speciation dynamics using Kraken, the Cray XT5
system at the National Institute for Computational Sciences. The problem's
underlying quadratic complexity was problematic, and the numerical
instabilities we faced would either compromise or else severely complicate
large-population simulations. We present lessons learned from the computational
challenges encountered, and describe how we have dealt with them
within the constraints presented by hardware.
|
Download:  cug2009.pdf cug2009.pdf
|
 Chapter: Dynamic patterns of adaptive radiation: evolution of mating preferences. Chapter: Dynamic patterns of adaptive radiation: evolution of mating preferences.
Dynamic patterns of adaptive radiation: evolution of mating preferences.
Gavrilets, S. and A. Vose
Chapter 7 of Speciation and Patterns of Diversity (2009), Cambridge University Press, pp. 102-126.
In Butlin, RK, J Bridle, and D Schluter (eds)
|
Adaptive radiation is defined as the evolution of ecological and phenotypic
diversity within a rapidly multiplying lineage (Simpson 1953; Schluter 2000).
Examples include the diversification of Darwin's finches on the Galapagos
islands, Anolis lizards on Caribbean islands, Hawaiian silverswords, a mainland
radiation of columbines, and cichlids of the East African Great Lakes, among
many others (Simpson 1953; Givnish & Sytsma 1997; Losos 1998; Schluter 2000;
Gillespie 2004; Salzburger & Meyer 2004; Seehausen 2006). Adaptive radiation
typically follows the colonization of a new environment or the establishment of
a ‘key innovation’ (e.g. nectar spurs in columbines, Hodges 1997) which opens
new ecological niches and/or new paths for evolution.
|
Download:  sniche.pdf sniche.pdf
|
 Paper: Palms on an Oceanic Island Paper: Palms on an Oceanic Island
Case studies and mathematical models of ecological speciation. 2. Palms on an oceanic island
Sergey Gavrilets and Aaron Vose
2007 Molecular Ecology, Blackwell Publishing Ltd
|
A recent study of a pair of sympatric species of palms on the Lord Howe Island is
viewed as providing probably one of the most convincing examples of sympatric speciation
to date. Here we describe and study a stochastic, individual-based, explicit genetic model
tailored for this palms system. Overall, our results show that relatively rapid (< 50 000
generations) colonization of a new ecological niche, and sympatric or parapatric speciation
via local adaptation and divergence in flowering periods are theoretically plausible if
(i) the number of loci controlling the ecological and flowering period traits is small; (ii) the
strength of selection for local adaptation is intermediate; and (iii) an acceleration of flow-
ering by a direct environmental effect associated with the new ecological niche is present.
We discuss patterns and time-scales of ecological speciation identified by our model, and
we highlight important parameters and features that need to be studied empirically in
order to provide information that can be used to improve the biological realism and power
of mathematical models of ecological speciation.
|
Download:  palms.pdf palms.pdf
|
 Paper: Cichlids in a Crater Lake Paper: Cichlids in a Crater Lake
Case studies and mathematical models of ecological speciation. 1. Cichlids in a crater lake
Sergey Gavrilets, Aaron Vose, Marta Barluenga, Walter Salzburger, and Axel Meyer
2007 Molecular Ecology, Blackwell Publishing Ltd
|
A recent study of a pair of sympatric species of cichlids in Lake Apoyo in Nicaragua is
viewed as providing probably one of the most convincing examples of sympatric speciation
to date. Here, we describe and study a stochastic, individual-based, explicit genetic model tai-
lored for this cichlid system. Our results show that relatively rapid (< 20 000 generations)
colonization of a new ecological niche and (sympatric or parapatric) speciation via local
adaptation and divergence in habitat and mating preferences are theoretically plausible if:
(i) the number of loci underlying the traits controlling local adaptation, and habitat and
mating preferences is small; (ii) the strength of selection for local adaptation is intermediate;
(iii) the carrying capacity of the population is intermediate; and (iv) the effects of the loci
influencing nonrandom mating are strong. We discuss patterns and timescales of ecological
speciation identified by our model, and we highlight important parameters and features
that need to be studied empirically to provide information that can be used to improve the
biological realism and power of mathematical models of ecological speciation.
|
Download:  cich.pdf cich.pdf
|
 Paper: The dynamics of Machiavellian intelligence Paper: The dynamics of Machiavellian intelligence
The dynamics of Machiavellian intelligence
Sergey Gavrilets and Aaron Vose
2006 Proc. Natl. Acad. Sci. USA 103: 16823-16828
|
The "Machiavellian intelligence" hypothesis (or the "social brain" hypothesis) posits that large brains and distinctive cognitive abilities of humans have evolved via intense social competition in which social competitors developed increasingly sophisticated "Machiavellian" strategies as a means to achieve higher social and reproductive success. Here we build a mathematical model aiming to explore this hypothesis. In the model, genes control brains which invent and learn strategies (memes) which are used by males to gain advantage in competition for mates. We show that the dynamics of intelligence has three distinct phases. During the dormant phase only newly invented memes are present in the population. During the cognitive explosion phase the population?s meme count and the learning ability, cerebral capacity (controlling the number of different memes that the brain can learn and use), and Machiavellian fitness of individuals increase in a runaway fashion. During the saturation phase natural selection resulting from the costs of having large brains checks further increases in cognitive abilities. Overall, our results suggest that the mechanisms underlying the "Machiavellian intelligence" hypothesis can indeed result in the evolution of significant cognitive abilities on the time scale of 10 to 20 thousand generations. We show that cerebral capacity evolves faster and to a larger degree than learning ability. Our model suggests that there may be a tendency toward a reduction in cognitive abilities (driven by the costs of having a large brain) as the reproductive advantage of having a large brain decreases and the exposure to memes increases in modern societies.
|
Download:  mi.pdf mi.pdf
|
 Paper: Dynamic Patterns of Adaptive Radiation Paper: Dynamic Patterns of Adaptive Radiation
Dynamic patterns of adaptive radiation
Sergey Gavrilets and Aaron Vose
2005 Proc. Natl. Acad. Sci. USA 102: 18040-18045
|
Adaptive radiation is defined as the evolution of ecological and phenotypic diversity within a rapidly multiplying lineage. When it occurs, adaptive radiation typically follows the colonization of a new environment or the establishment of a "key innovation," which opens new ecological niches andor new paths for evolution. Here, we take advantage of recent developments in speciation theory and modern computing power to build and explore a large-scale, stochastic, spatially explicit, individual-based model of adaptive radiation driven by adaptation to multidimensional ecological niches. We are able to model evolutionary dynamics of populations with hundreds of thousands of sexual diploid individuals over a time span of 100,000 generations assuming realistic mutation rates and allowing for genetic variation in a large number of both selected and neutral loci. Our results provide theoretical support and explanation for a number of empirical patterns including "area effect," "overshooting effect," and "least action effect," as well as for the idea of a "porous genome." Our findings suggest that the genetic architecture of traits involved in the most spectacular radiations might be rather simple. We show that a great majority of speciation events are concentrated early in the phylogeny. Our results emphasize the importance of ecological opportunity and genetic constraints in controlling the dynamics of adaptive radiation.
|
Download:  ar.pdf ar.pdf
|
 | Old Stuff: |
- I have a page of
 images, images,  videos, and videos, and  interesting links in the "Pile". interesting links in the "Pile".
-
 QRGA:
Fancy QR codes via genetic algorithms. Work in progress; check out it's dedicated page: QRGA and at GitHub: QRGA. QRGA:
Fancy QR codes via genetic algorithms. Work in progress; check out it's dedicated page: QRGA and at GitHub: QRGA.
-
 CardCloud:
A web-based interface where players can manipulate cards. Work in progress; check out it's dedicated page: CardCloud and at GitHub: CardCloud. CardCloud:
A web-based interface where players can manipulate cards. Work in progress; check out it's dedicated page: CardCloud and at GitHub: CardCloud.
-
 ChromoCraft:
This is a project I've been working on from time to time just for fun.
ChromoCraft is a 3D tower defense game for linux with some neat features like sound, etc.
Check out the page I've made for it with complete source code here. ChromoCraft:
This is a project I've been working on from time to time just for fun.
ChromoCraft is a 3D tower defense game for linux with some neat features like sound, etc.
Check out the page I've made for it with complete source code here.
-
 iSmartMailboxHD.com:
An Arduino-based smart mailbox project, for fun. Check out it's dedicated page: iSmartMailboxHD.com (ismbhddc/). iSmartMailboxHD.com:
An Arduino-based smart mailbox project, for fun. Check out it's dedicated page: iSmartMailboxHD.com (ismbhddc/).
-
 PhyTree:
I'm working on a toolchain that turns raw protein sequences into a phylogenetic tree / tree of life,
as well as web-based viewing software to make the data accessible. My preferred layout algorithm for
the tree is force-directed placement. I'm working on 2D and 3D versions:
For the 3D version, I have an alpha using WebGL
(as well as more advanced CUDA and OpenCL versions that need work).
For the 2D version, I have an alpha using browser-based SVG
(Scalable Vector Graphics). Currently, I recommend using Google Chrome to view these pages as it
has the best performance I have seen to date. PhyTree:
I'm working on a toolchain that turns raw protein sequences into a phylogenetic tree / tree of life,
as well as web-based viewing software to make the data accessible. My preferred layout algorithm for
the tree is force-directed placement. I'm working on 2D and 3D versions:
For the 3D version, I have an alpha using WebGL
(as well as more advanced CUDA and OpenCL versions that need work).
For the 2D version, I have an alpha using browser-based SVG
(Scalable Vector Graphics). Currently, I recommend using Google Chrome to view these pages as it
has the best performance I have seen to date.
-
 fsmem:
I created a software library which allows applications to use space on
disk instead of RAM for malloc()-based memory. While swap space in
Linux is nice, it is very uncommon to create an install with a large
(think terabytes) swap partition. Additionally, one might not want to
dedicate that much disk space to swap. The fsmem
library allows one to enable a specific application to use files on
disk for memory instead. This is done by memory mapping large files
and then allowing ptmalloc to
carve them up transparently. fsmem:
I created a software library which allows applications to use space on
disk instead of RAM for malloc()-based memory. While swap space in
Linux is nice, it is very uncommon to create an install with a large
(think terabytes) swap partition. Additionally, one might not want to
dedicate that much disk space to swap. The fsmem
library allows one to enable a specific application to use files on
disk for memory instead. This is done by memory mapping large files
and then allowing ptmalloc to
carve them up transparently.
-
 Cellular
Automata: Check out the dedicated page for
the CA simulator and the related Hopfield neural network project here. Cellular
Automata: Check out the dedicated page for
the CA simulator and the related Hopfield neural network project here.
-
 Peptide
MW Calculator: This allows input of protein sequences in FASTA format and outputs the monoisotopic and averageisotopic molecular weight: peptide_Mar-19-2013.tar.gz. Peptide
MW Calculator: This allows input of protein sequences in FASTA format and outputs the monoisotopic and averageisotopic molecular weight: peptide_Mar-19-2013.tar.gz.
 Very Old Projects: There are some extremely old projects provided at the very bottom of this page. Very Old Projects: There are some extremely old projects provided at the very bottom of this page.
 |
The following projects, while perhaps interesting or fun, are quite a few years old now and seem a bit hackish by current standards.
Please understand that they are provided along with some embarrassment.
|
 pTree: Process Tree Viewer pTree: Process Tree Viewer
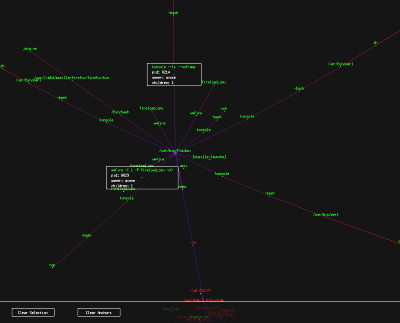
|
I was inspired by `ps auxf`.
If ps can display an ascii tree, I can certainly do better in
OpenGL.
pTree parses the
output of `ps -ef` to gain information about currently
running processes and uses that information to build a process tree in
memory. The process tree is then presented in a fully interactive
3D environment while a background thread applies anti-gravity and
spring physics to cause the tree to self organize in space. This technique
is commonly called "force-directed placement".
Red processes are owned by root, green processes are owned by the
current user, and orange processes are owned by someone else. The
edge color is related its depth in the process tree.
This is still a work in progress and I plan many interface
enhancements. With that in mind, I'll tell you some of the
controls to get you started:
Left mouse down:
Left mouse drag:
Right mouse drag:
Mouse Wheel:
Middle mouse down:
Middle mouse drag:
|
Toggle Select
Move Tree X Y
Rotate Tree X Y Z
Move Tree Z
Toggle Anchor
Drag Node
|
Download:  ptree.tar.bz2 ptree.tar.bz2
|
 mChat: 3D Chat Server/Client mChat: 3D Chat Server/Client
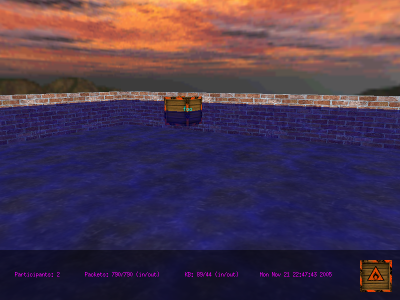
|
This is a client/server application
that allows the participants to "swim" in a virtual pool floating in
the middle of the air. The participants can't actually chat with
one another, but my excuse is that this is mostly a demonstration
program. Still,
it's fun to play with.
mChat adds a layer of checksums on top of what UDP provides, shows a
working skybox implementation, and is multi-threaded.
The the standard "wasd" system is used to move, and dragging with the
left mouse button will look around (high sensitivity). The right
mouse button will cycle through a short list of avatar skins (your skin
resembles the sample in the bottom right corner).
Download:  mchat.tar.bz2 mchat.tar.bz2
|
|

 Resume / Curriculum Vitae.
Resume / Curriculum Vitae.
 Paper: Evaluating Gather and Scatter Performance on CPUs and GPUs
Paper: Evaluating Gather and Scatter Performance on CPUs and GPUs Paper: PharML.Bind: Pharmacologic Machine Learning for Protein-Ligand Interactions
Paper: PharML.Bind: Pharmacologic Machine Learning for Protein-Ligand Interactions Paper: Recombination of Artificial Neural Networks
Paper: Recombination of Artificial Neural Networks Paper: Interactive Distributed Deep Learning with Jupyter Notebooks
Paper: Interactive Distributed Deep Learning with Jupyter Notebooks Cray Blog: How to Program a Supercomputer (October 24, 2017)
Cray Blog: How to Program a Supercomputer (October 24, 2017) Cray Blog: The Intersection of Machine Learning and High-Performance Computing (August 31, 2017)
Cray Blog: The Intersection of Machine Learning and High-Performance Computing (August 31, 2017) Paper: An MPI/OpenACC implementation of a high-order electromagnetics solver with GPUDirect communication.
Paper: An MPI/OpenACC implementation of a high-order electromagnetics solver with GPUDirect communication. Paper: Massively parallel and linear-scaling algorithm for second-order Moller-Plesset perturbation theory applied to the study of supramolecular wires.
Paper: Massively parallel and linear-scaling algorithm for second-order Moller-Plesset perturbation theory applied to the study of supramolecular wires. Paper: Relative debugging for a highly parallel hybrid computer system.
Paper: Relative debugging for a highly parallel hybrid computer system. Paper: A case study of CUDA FORTRAN and OpenACC for an atmospheric climate kernel.
Paper: A case study of CUDA FORTRAN and OpenACC for an atmospheric climate kernel. Cray Blog: HPC Software Optimization Trends (Part 1): Where Did We Come From and Where Are We Now? (August 11, 2015)
Cray Blog: HPC Software Optimization Trends (Part 1): Where Did We Come From and Where Are We Now? (August 11, 2015) Cray Blog: HPC Software Optimization Trends (Part 2): What Will the Future Be Like and How Do We Prepare for It? (August 12, 2015)
Cray Blog: HPC Software Optimization Trends (Part 2): What Will the Future Be Like and How Do We Prepare for It? (August 12, 2015) Paper: Tri-Hybrid Computational Fluid Dynamics on DoE's Cray XK7, Titan.
Paper: Tri-Hybrid Computational Fluid Dynamics on DoE's Cray XK7, Titan. Paper: HSPp-BLAST: Highly Scalable Parallel PSI-BLAST for Very Large-scale Sequence Searches
Paper: HSPp-BLAST: Highly Scalable Parallel PSI-BLAST for Very Large-scale Sequence Searches Paper: Patterns of Species Ranges, Speciation, and Extinction
Paper: Patterns of Species Ranges, Speciation, and Extinction Paper: A Pragmatic Approach to Improving the Large-scale Parallel I/O Performance of Scientific Applications.
Paper: A Pragmatic Approach to Improving the Large-scale Parallel I/O Performance of Scientific Applications. Paper: Simulating Population Genetics on the XT5
Paper: Simulating Population Genetics on the XT5 Chapter: Dynamic patterns of adaptive radiation: evolution of mating preferences.
Chapter: Dynamic patterns of adaptive radiation: evolution of mating preferences. Paper: Palms on an Oceanic Island
Paper: Palms on an Oceanic Island Paper: Cichlids in a Crater Lake
Paper: Cichlids in a Crater Lake Paper: The dynamics of Machiavellian intelligence
Paper: The dynamics of Machiavellian intelligence Paper: Dynamic Patterns of Adaptive Radiation
Paper: Dynamic Patterns of Adaptive Radiation pTree: Process Tree Viewer
pTree: Process Tree Viewer mChat: 3D Chat Server/Client
mChat: 3D Chat Server/Client Sokoban: One more Sokoban clone
Sokoban: One more Sokoban clone





 images,
images,  videos, and
videos, and  interesting links in the "
interesting links in the "